Accelerating Vaccine Development with Transcriptomics
The landscape of vaccine development is undergoing a significant transformation with the advent of transcriptomic approaches.
These technologies offer unprecedented insights into the complexities of the immune system, enabling the creation of more effective and targeted vaccines.
Understanding Transcriptomics
In the cutting-edge field of vaccine development, transcriptomics stands out as a revolutionary approach, offering unprecedented insights into the intricate molecular mechanisms underlying immune responses. But what exactly is transcriptomics, and why is it so crucial for advancing vaccine research?
What are transcriptomics and why are they important?
Transcriptomics is the comprehensive study of the transcriptome—the complete set of RNA molecules, or transcripts, expressed in a particular cell, tissue, or organism at any given time.1 Unlike genomics, which focuses on the static DNA blueprint, transcriptomics captures the dynamic expression patterns of genes, providing a snapshot of how genetic information is being utilized by cells in response to various conditions.2 By employing advanced high-throughput sequencing technologies and sophisticated bioinformatics tools, researchers can analyze the complete set of RNA transcripts produced by the genome, revealing how cells respond to stimuli, such as infections or vaccinations.
Two primary techniques dominate the field of transcriptomics: microarrays and RNA sequencing (RNA-Seq). Microarrays quantify a predefined set of RNA sequences, while RNA-Seq employs high-throughput sequencing to capture all RNA sequences present in a sample. This comprehensive approach allows for a more detailed understanding of gene expression changes.3
In vaccine development, transcriptomics provides comprehensive insights into the molecular mechanisms underlying immune responses to vaccination. This capability allows researchers to identify transcriptomic signatures associated with protective immune responses, serving as correlates of protection for vaccine efficacy.4 Furthermore, understanding these molecular mechanisms aids in the rational design of vaccines, enabling scientists to engineer vaccines that elicit the most effective immune responses by targeting specific genes and pathways identified through transcriptomic studies.5
Transcriptomics also facilitates the comparison of different vaccine formulations and adjuvants, optimizing vaccine composition for better efficacy and safety.5 Additionally, gene expression profiles can predict the potential efficacy of vaccines, allowing for more efficient development and testing.4 The analysis of transcriptomic data reveals the activation of innate immune pathways, which are essential for initiating adaptive immune responses, thus informing the design of vaccines that effectively stimulate both aspects of immunity.6
This approach also aids in identifying novel genes and pathways involved in vaccine-induced immunity, uncovering new targets for vaccine development. Monitoring changes in gene expression contributes to safety assessments, ensuring vaccines are not only effective but also safe for widespread use.7
Integrating transcriptomics with other “omics” data, such as proteomics and metabolomics, provides a holistic understanding of vaccine-induced immune responses, uncovering complex interactions between different molecular components.
Moreover, transcriptomics paves the way for personalized vaccination strategies by analyzing individual transcriptomic responses, potentially leading to vaccines tailored to an individual’s genetic makeup for optimized effectiveness and minimized adverse effects.7
In essence, transcriptomics revolutionizes vaccine development by offering detailed insights into immune responses, guiding the design of more effective and safer vaccines.
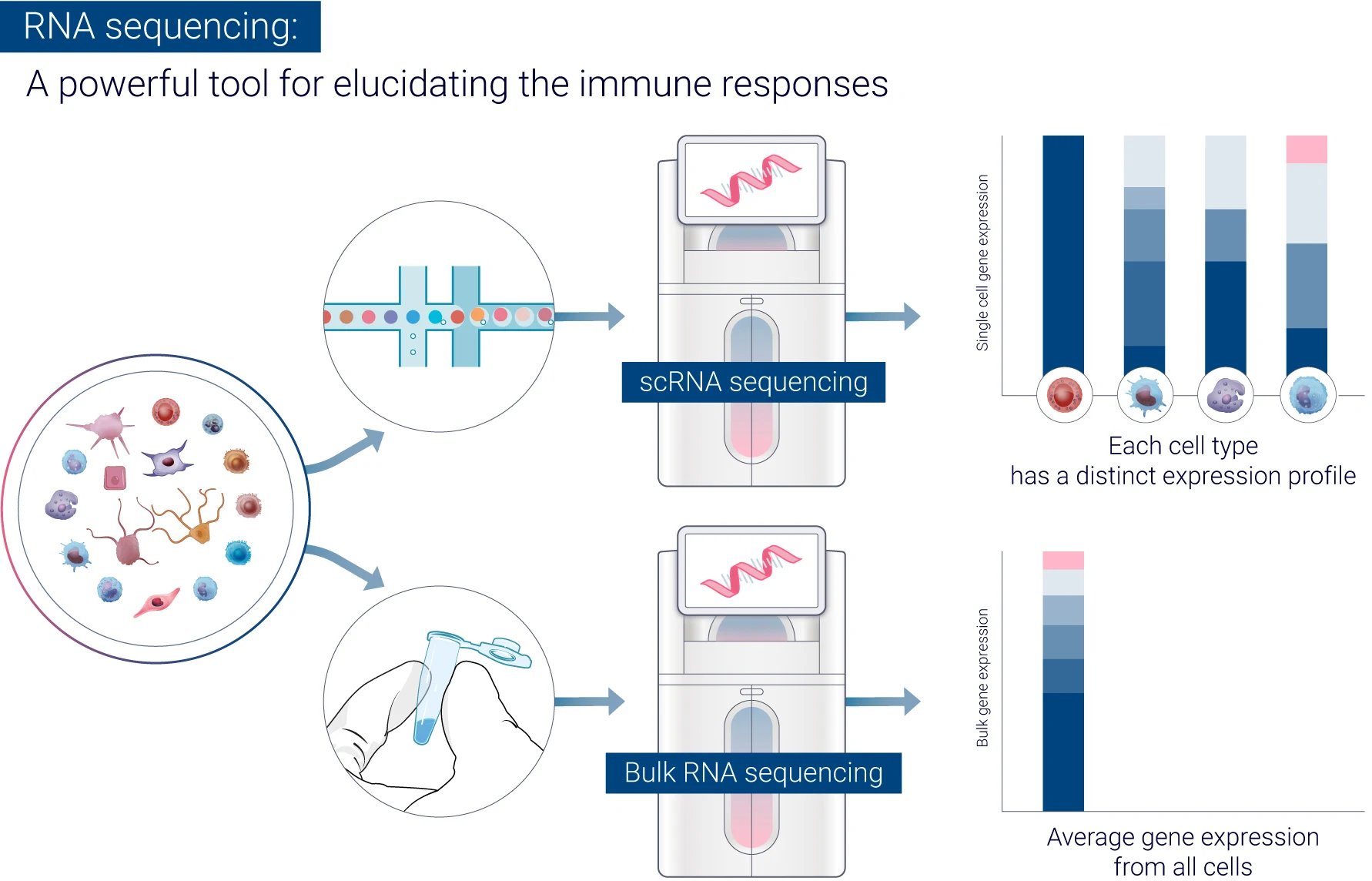
How can RNA sequencing provide insights into immune responses?
RNA-seq has emerged as a powerful tool for elucidating the immune responses triggered by vaccine injections, providing several key insights into these complex biological processes. One of its major contributions is the characterization of adaptive immunity, where RNA-seq can detect the activation and differentiation of T cells and B cells by analyzing changes in gene expression related to these immune processes.8 Additionally, RNA-seq captures the temporal dynamics of the immune response by sampling at multiple time points, allowing researchers to observe how gene expression evolves over hours, days, and weeks post-vaccination.
The ability to profile cell type-specific responses through single-cell RNA-seq further enhances our understanding by revealing how individual immune cell types respond to vaccines. This granularity is essential for comparing immune signatures induced by different vaccine platforms, such as mRNA vaccines versus protein subunit vaccines.9 Moreover, RNA-seq facilitates the correlation of gene expression patterns with antibody responses, helping to identify potential transcriptional biomarkers of vaccine efficacy.
Another significant insight provided by RNA-seq is the detection of inflammatory responses, as it reveals the induction of inflammatory pathways that may be related to reactogenicity, or vaccine side effects. The technology also uncovers previously unknown genes involved in vaccine-induced immunity, expanding the pool of potential targets for future vaccine development. On a systems level, RNA-seq offers a global perspective on the coordinated immune response, elucidating gene networks and pathways activated by vaccination.
By providing these comprehensive insights, RNA-seq has become indispensable for understanding vaccine-induced immunity and guiding the development strategies and optimization of vaccine efficacy.
Transcriptomics Impact on Vaccine Development
Case study: Transcriptomics reveals distinct immune signatures of SARS-CoV-2 Vaccines
In a study published in Emerging Microbes and Infections, Wang et al. compared the innate immune responses elicited by two SARS-CoV-2 vaccines: the protein subunit vaccine ZF2001 and the mRNA vaccine RRV.8 Despite both showing high protective efficacy in clinical trials, their mechanisms of action were not fully understood. Using transcriptome sequencing, the study analyzed gene expression patterns in mice at multiple time points after immunization, offering a comprehensive, time-dependent analysis of immune response signatures.
The findings revealed distinct patterns of gene activation between the two vaccines. ZF2001 quickly induced MHC class II-related genes, while RRV more rapidly induced MHC class I-related genes. RRV also demonstrated a robust early response, including activation of pattern recognition receptors and genes involved in RNA degradation and transcription inhibition. Notably, gene expression patterns at day 7 correlated with antibody titers at day 21, suggesting potential early markers of vaccine efficacy.
This study highlighted several benefits of transcriptomics: it provided a comprehensive analysis of thousands of genes, revealed the dynamics of gene expression over time, and offered mechanistic insights into specific genes and pathways activated by each vaccine. The ability to correlate early gene expression with later antibody responses suggests potential biomarkers for vaccine efficacy. Moreover, transcriptomics enabled a direct comparison between the two vaccine types, identifying their unique and shared characteristics. This case study underscores the value of transcriptomics in understanding vaccine-induced immune responses and guiding the development and optimization of future vaccines.
Impact on public health and disease prevention
Transcriptomics has had a significant impact on vaccine development, with important implications for public health and disease prevention.
Here are some of the key impacts:
- Personalized vaccination strategies: Transcriptomics reveals individual variations in immune responses, potentially leading to more personalized vaccination approaches that consider factors like age, genetics, and pre-existing immunity. Adjuvants, doses, vaccination schedules, and the need for a booster can be tailored to different individuals or groups thanks to the new understanding of vaccine response through transcriptomics.7
- Rapid response to emerging threats: Transcriptomics enables the understanding of pathogen biology and host-pathogen interactions, helping to identify the correct targets and to design the most effective vaccine in a shorter time frame. Transcriptomics also facilitates fast-track candidate selection by revealing which antigens elicit strong immune responses. It also allows for real-time monitoring, providing data to refine and improve vaccines on the fly during outbreaks.6
- Identification of novel targets: Transcriptomic studies can uncover previously unknown genes and pathways involved in vaccine-induced immunity, potentially leading to new vaccine targets and strategies such as broad-spectrum vaccines or mRNA vaccines.5
- Optimization of vaccination schedules: Analysis of temporal dynamics of immune responses helps in understanding the optimal timing for boosters to sustain immunity. Gene expression changes can guide the development of combination vaccines and their schedules, ensuring that multiple antigens are presented in a way that elicits the strongest and most durable immune response.4
By leveraging these insights from transcriptomics, vaccine development can become more precise, efficient, and tailored to the needs of diverse populations, ultimately enhancing public health and disease prevention efforts.
Challenges and Future Directions
Current limitations and ongoing research
While transcriptomics holds great promise for advancing vaccine development, several key challenges remain.
The complexity of transcriptomic data, involving thousands of genes, makes data interpretation difficult. Ongoing research focuses on advanced bioinformatics and machine learning to address this. Also, researchers are working on integrating transcriptomics with other “omics” data (proteomics, metabolomics, etc.) and immunological readouts to provide a more comprehensive understanding of vaccine-induced immunity.10 Additionally, there is ongoing work to establish standardized protocols and quality control measures to improve the reproducibility of transcriptomic results across different laboratories and studies. Regarding data sharing and meta-analysis, efforts are underway to improve it across multiple vaccine studies and identify common protective signatures.
Additionally, many transcriptomics studies are still conducted in animal models. A major challenge is translating these findings to human vaccine responses due to species differences. More human studies and improved models that better recapitulate human immune responses are needed. Also, small sample sizes in many studies limit statistical power. Determining optimal sample sizes and sequencing depths is an ongoing effort.
Regarding the outcomes of transcriptomics for vaccine research, differentiating vaccine-specific transcriptional changes from general inflammatory responses remains challenging. More research is needed to identify vaccine-specific signatures. While transcriptomics can identify immune signatures, correlating these with actual protection against disease remains difficult. Long-term studies correlating early transcriptional changes with clinical outcomes are needed.
Finally, although costs have decreased, RNA-seq remains expensive for large-scale vaccine trials. Research into more cost-effective approaches is ongoing.
Addressing these challenges is essential for fully leveraging transcriptomics in vaccine development, translating insights into better vaccine design and evaluation strategies.
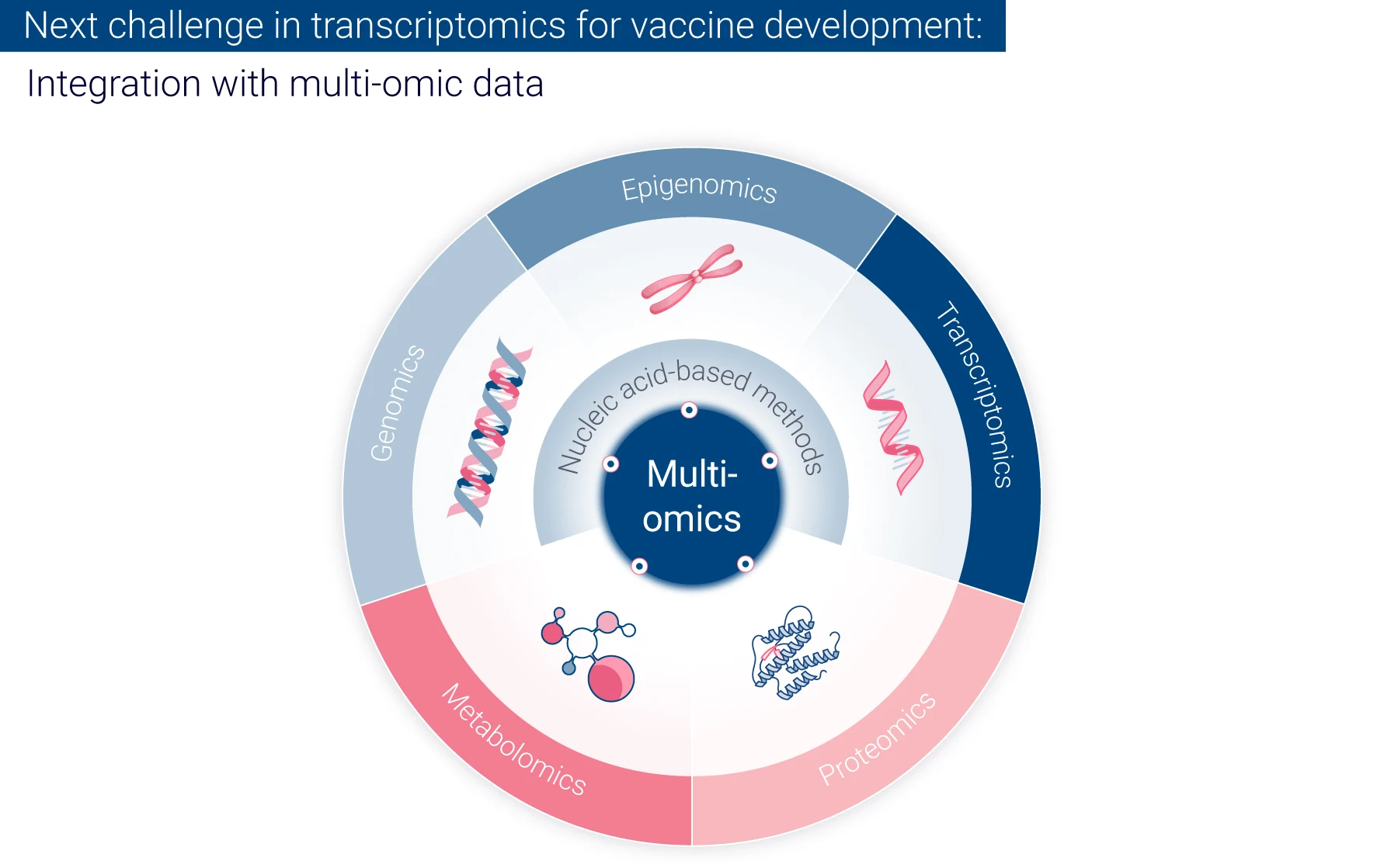
The future potential of transcriptomics for vaccine development
The future potential of transcriptomics in vaccine development is significant and promising. Here are some key areas where transcriptomics is expected to have a major impact11:
- Advanced bioinformatics and machine learning: The development of more sophisticated bioinformatics tools and machine learning algorithms will enhance our ability to extract meaningful insights from complex transcriptomic data, leading to better identification of correlates of protection and biomarkers of vaccine efficacy.
- Integration with other omics-based data: Future research will likely focus on integrating transcriptomics with other omics data (e.g., proteomics, metabolomics) to provide a more comprehensive understanding of vaccine-induced immunity.
- Single-cell and spatial transcriptomic: Advancements in single-cell RNA-seq and spatial transcriptomics technologies will allow for more detailed analysis of cell type-specific responses and tissue-specific immune reactions to vaccines.
- Cost reduction and accessibility: As with other genomic technologies, the cost of transcriptomics is expected to decrease, making it more accessible for routine use in vaccine development and clinical trials.
Concluding Thoughts
By leveraging the power of transcriptomic approaches, researchers are paving the way for a new era in vaccine development, one that promises to be more precise, effective, and responsive to global health challenges.
References
1Rao S, Ghosh D, Asturias EJ, Weinberg A. What can we learn about influenza infection and vaccination from transcriptomics? Hum Vaccin Immunother. 2019;15(11):2615-2623. doi: 10.1080/21645515.2019.1608744. Epub 2019 May 22. PMID: 31116679; PMCID: PMC6930070.
2Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009 Jan;10(1):57-63. doi: 10.1038/nrg2484. PMID: 19015660; PMCID: PMC2949280.
3Lowe R, Shirley N, Bleackley M, Dolan S, Shafee T. Transcriptomics technologies. PLoS Comput Biol. 2017 May 18;13(5):e1005457. doi: 10.1371/journal.pcbi.1005457. PMID: 28545146; PMCID: PMC5436640.
4Barton AJ, Hill J, Pollars AJ, Blohmke CJ. Transcriptomics in Human Challenge Models. Front. Immunol. 2017 Dec; 8:1664-3224. doi:10.3389/fimmu.2017.01839
5Santoro F, Pettini E, Kazmin D, Ciabattini A, Fiorino F, Gilfillan GD, Evenroed IM, Andersen P, Pozzi G and Medaglini D (2018) Transcriptomics of the Vaccine Immune Response: Priming With Adjuvant Modulates Recall Innate Responses After Boosting. Front. Immunol. 9:1248. doi: 10.3389/fimmu.2018.01248
6Zhang Y, Guo X, Li C, Kou Z, Lin L, Yao M, Pang B, Zhang X, Duan Q, Tian X, Xing Y, Jiang X. Transcriptome Analysis of Peripheral Blood Mononuclear Cells in SARS-CoV-2 Naïve and Recovered Individuals Vaccinated With Inactivated Vaccine. Front Cell Infect Microbiol. 2022 Feb 3;11:821828. doi: 10.3389/fcimb.2021.821828. PMID: 35186784; PMCID: PMC8851474.
7Voigt, E.A., Grill, D.E., Zimmermann, M.T. et al. Transcriptomic signatures of cellular and humoral immune responses in older adults after seasonal influenza vaccination identified by data-driven clustering. Sci Rep 8, 739 (2018). https://doi.org/10.1038/s41598-017-17735-x
8Wang Q, Song Z, Yang J, He Q, Mao Q, Bai Y, Liu J, An C, Yan X, Cui B, Song L, Liu D, Xu M, Liang Z. Transcriptomic analysis of the innate immune signatures of a SARS-CoV-2 protein subunit vaccine ZF2001 and an mRNA vaccine RRV. Emerg Microbes Infect. 2022 Dec;11(1):1145-1153. doi: 10.1080/22221751.2022.2059404. PMID: 35343384; PMCID: PMC9037177.
9Park, HJ., Bang, YJ., Kwon, S.P. et al. Analyzing immune responses to varied mRNA and protein vaccine sequences. npj Vaccines 8, 84 (2023). https://doi.org/10.1038/s41541-023-00684-0
10Goll JB, Bosinger SE, Jensen TL, Walum H, Grimes T, Tharp GK, Natrajan MS, Blazevic A, Head RD, Gelber CE, Steenbergen KJ, Patel NB, Sanz P, Rouphael NG, Anderson EJ, Mulligan MJ and Hoft DF (2023) The Vacc-SeqQC project: Benchmarking RNA-Seq for clinical vaccine studies. Front. Immunol. 13:1093242. doi: 10.3389/fimmu.2022.1093242
11Martínez-Pérez A, Estévez O, González-Fernández Á. Contribution and Future of High-Throughput Transcriptomics in Battling Tuberculosis. Front Microbiol. 2022 Feb 24;13:835620. doi: 10.3389/fmicb.2022.835620. PMID: 35283833; PMCID: PMC8908424.
Comments are closed.



