Redefining Local Toxicity Assessment
Part 2 – The Role of Animal-Free Non-Clinical Platforms in Drug Development.
In the realm of non-clinical animal-free platforms, a transformative shift is unfolding in response to the challenges posed by drug-induced adverse reactions
a predominant factor leading to the discontinuation of drug development projects and the withdrawal of drugs from the market1.
The imperative for accurate, ethical, and human-relevant methods to assess the toxicity and tolerance of new drug candidates has never been more crucial. Traditional animal testing methods come with inherent limitations, including ethical concerns and species-specific differences that may not accurately reflect human responses.
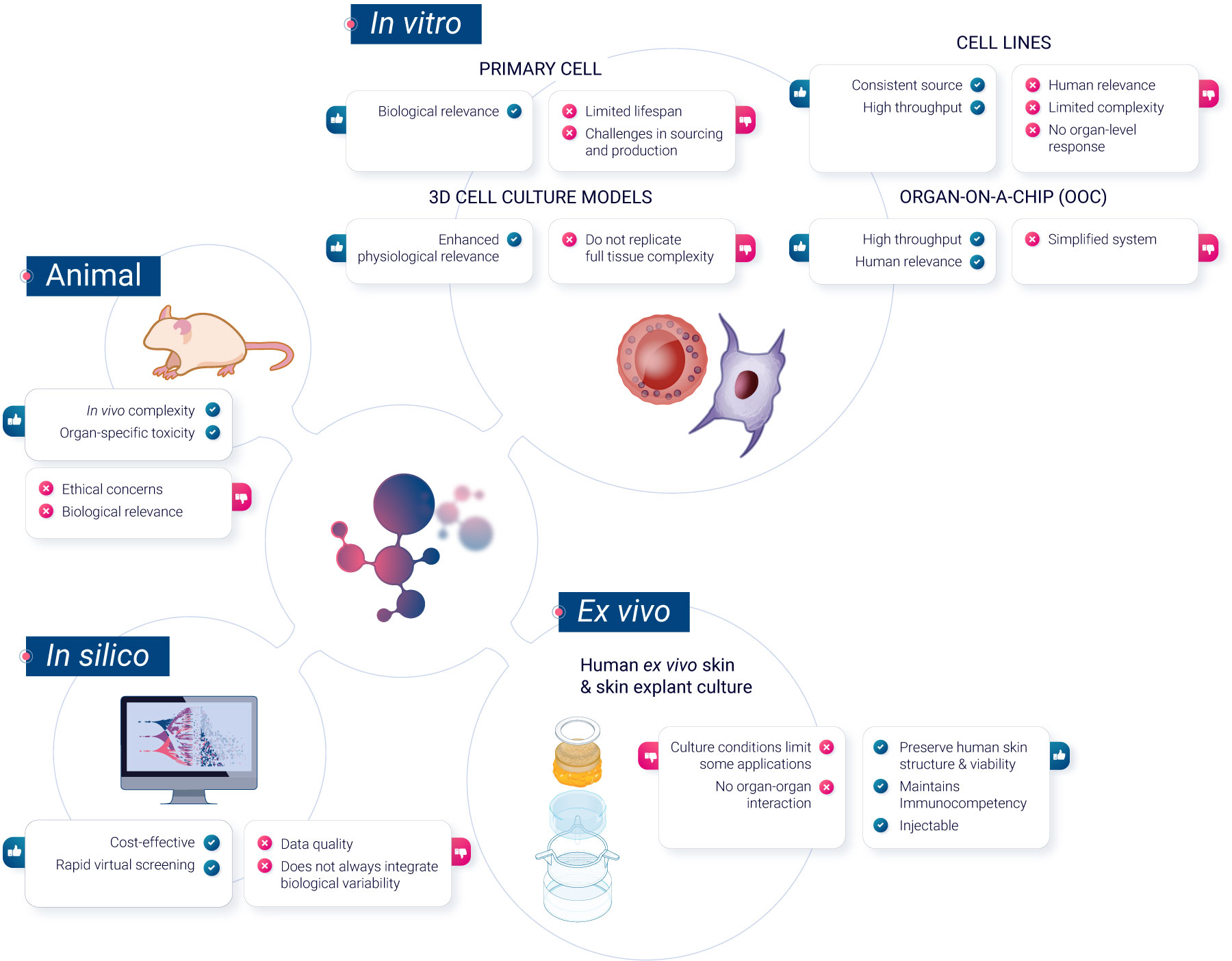
To navigate these challenges, scientists have developed a range of non-animal methods, each offering unique advantages. In this article, we will explore these methods, grouped for clarity into 3 categories: in vitro cell culture models, ex vivo models, and computer-based predictive models.
In vitro methods
Primary cells
Primary cells, sourced or developed in vitro from blood or immune organs, offer a more realistic representation of in vivo cell reactivity than immortalized cell lines. They do not undergo any modifications, unlike cell lines. Primary cells maintain the physiological properties and functions of the tissue from which they were derived. They have a limited lifespan in culture and cannot be propagated. Primary cells are also generally more representative of how cells in the body respond to drugs, making them more suitable for drug development and toxicity testing.
However, they present challenges, including sourcing difficulties and response variability between donors. While this variability is reduced in human innate immunity, assessing immunosafety in primary cells demands testing multiple donors to discern therapeutic compound effects from population response variations.
Sourcing remains a limitation, with most assessments focusing on easily accessible cells, missing organ-specific immune reactions.
Cell lines
Classical in vitro systems primarily use immortalized cell lines, which are derived from tumor cells or transformed cells capable of unlimited divisions in the absence of cellular senescence. They provide a consistent source with uniform characteristics. These cell lines can be genetically altered to study specific gene functions, aiding in analyzing interactions with nanomaterials in immune cells. Numerous human, mouse, and insect cell lines representing various immune cells are available. However, their representation of primary cells is questionable and must be validated for each biomarker2.
3D cell models
3D cell culture represents a shift from traditional 2D cell cultures by allowing cells to grow in three dimensions, more closely mimicking the natural environment of tissues in the body. These models offer a more physiologically-relevant context for studying cell behavior, interactions, and responses to stimuli3. However, they may not always replicate the full complexity of in vivo conditions. Examples of 3D cell culture models include spheroïds, which are spherical aggregates of cells, and organoïds, miniaturized and simplified versions of organs. While 3D cell culture models can provide valuable insights in various research areas, they are one of many tools available, each with its specific advantages and challenges in biomedical research.
Reconstructed human epidermis
Reconstructed Human Epidermis (RHE) is a laboratory-developed model designed to resemble the human epidermal structure. It is primarily used for topical toxicity testing and other applications that require an understanding of epidermal responses. While RHE provides a representation of the human epidermis, it is limited in its ability to capture systemic effects or deeper skin layers. As a result, RHE serves as one of several models in the research toolkit, each with its specific applications and limitations in the field of dermatological and pharmaceutical research.
Organoïds
Organoïds are advanced 3D culture systems whose structure can closely mimic organ development and various human diseases, including infectious diseases, genetic abnormalities, and cancers. Organoïds can be derived from various sources, including embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), adult stem cells, and even primary tumor cells.
Skin organoïds are usually derived from iPSCs. They have been successfully developed for multiple organs, such as the lung, stomach, intestine, liver, pancreas, prostate, and brain. Despite their potential, there are still limitations to organoïd technology that need to be addressed such as the lack of vascular system or immune system network. Organoïds may not fully recapitulate the entire tissue complexity, and some cell types or functions may be absent or underrepresented.
Organ-on-a-Chip (OoC)
Organ-on-a-Chip is a bioengineered microdevice that aims to replicate the functions of human organs and tissues within a controlled environment. Through the use of microfluidic channels, it facilitates tissue perfusion and provides an avenue for monitoring cell health and activity in real-time. While the technology offers a different approach to studying human physiological responses, there are complexities associated with replicating intricate organ-organ interactions5.
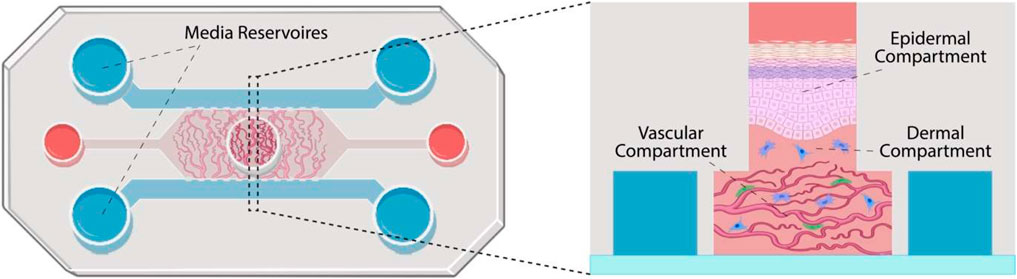
Ex vivo methods
Human ex vivo skin and skin explant culture have been traditionally used in investigative dermatology to study the physiology and pathophysiology of human skin and its epidermal appendages in vitro. Additionally, human ex vivo skin is recognized as a preferred tissue for regulatory dermal absorption experiments. However, the conditions under which human skin is maintained in culture have limited its applicability.
It’s crucial to maintain the physiological tissue viability when using human ex vivo skin, as evidenced by the degradation of steroidogenic enzymes during cryopreservation6.
Genoskin has developed a technology to biostabilize human skin explants for approximately seven days.
This system not only maintains viability of the skin structure, but also its immunocompetency. We have further advanced the development of these ready-to-use skin models, which can now be used as a proxy for studying the human immune system, enabling the prediction of the toxicity of pharmaceutical compounds administered through subcutaneous injection or topical and systemic application.
These capabilities elevate ex vivo skin models from investigative dermatology into the realm of predictive assessments for immunotoxicity and cytotoxicity.
In silico methods
In silico techniques have become the standard in drug discovery, and their potential benefits extend throughout the entire drug development process. For instance, they have been used since 1970 for the prediction of toxicity7 and methods such as computer-based predictive models, particularly quantitative structure-activity relationship (QSAR) models and machine learning algorithms, have gained prominence in toxicity prediction. These models utilize chemical and structural information to predict compound toxicity, making them invaluable in early-stage drug development.
QSAR models can predict the toxicity of compounds based on their chemical characteristics, offering a rapid and cost-effective screening tool. Machine learning models, trained on extensive datasets of chemical and biological information, can provide accurate predictions of compound toxicity and tolerance, aiding in the selection and prioritization of drug candidates.
Other methods such as 3D modeling of binding activities, modeling pharmacokinetics, models for repeated-dose toxicity, and others are regularly being developed and reviewed by Regulatory Agencies8.
Concluding Remark
We’ve journeyed through a world of non-clinical animal-free testing methods in drug development, each method bringing something unique to the table in understanding drug safety and effects. From cells cultured in petri dishes to computational algorithms predicting drug reactions – The goal is to ensure that research becomes more ethical and directly applicable to human concerns.
Next up in this series, we are diving into something groundbreaking! Imagine using ex vivo platforms for the very first steps in human clinical trials! Stay connected, as we explore how these platforms have the potential to significantly disrupt the traditional approaches of conducting initial clinical trials.
References
1Cook, David et al. “Lessons learned from the fate of AstraZeneca’s drug pipeline: a five-dimensional framework.” Nature reviews. Drug discovery vol. 13,6 (2014): 419-31.
2Boraschi, Diana et al. “In Vitro and In Vivo Models to Assess the Immune-Related Effects of Nanomaterials.” International journal of environmental research and public health vol. 18,22 11769. 10 Nov. 2021
3Weiskirchen, Sabine et al. “A Beginner’s Guide to Cell Culture: Practical Advice for Preventing Needless Problems.” Cells vol. 12,5 682. 21 Feb. 2023, doi:10.3390/cells12050682
4Front. Genet., 01 November 2021. Sec. Toxicogenomics Volume 12 – 2021 |Human Organoids for Predictive Toxicology Research and Drug Development
5 Leung, C.M., de Haan, P., Ronaldson-Bouchard, K. et al. A guide to the organ-on-a-chip. Nat Rev Methods Primers 2, 33 (2022).
6De Wever, B., Kurdylowski, S., Descargues, P. (2015) ‘Human Skin Models for Research Applications in Pharmacology and Toxicology: Introducing NativeSkin®, the “Missing Link” Bridging Cell Culture and/or Reconstructed Skin Models and Human Clinical Testing‘, Appl In Vitro Toxicol
7Amberg, A. (2013). In Silico Methods. In: Vogel, H.G., Maas, J., Hock, F.J., Mayer, D. (eds) Drug Discovery and Evaluation: Safety and Pharmacokinetic Assays. Springer, Berlin, Heidelberg.
8Benfenati, E.(2016). In Silico Methods for Predicting Drug Toxicity.
Comments are closed.